PRO-Seq
PRO-Seq
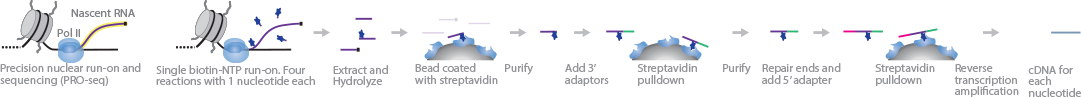
Precision nuclear run-on sequencing (PRO-Seq) maps RNA polymerase II pause sites with base-pair resolution during RNA transcription. This approach is similar to GRO-Seq, but provides the added benefit of single-base resolution. RNA pol II initiation sites can be mapped using a modified protocol named PRO-cap. In PRO-seq, four separate run-on reactions, each with only one type of biotin-NTP and sarkosyl, are carried out on nucleic lysates. Incorporation of the single biotin-NTP halts further elongation of nascent RNA strands by RNA pol II. The RNA strands are extracted, fragmented, and purified through streptavidin pull-down. 3' adaptors are directly ligated to the purified sample prior to another streptavidin purification step. 5' ends are repaired using tobacco acid pyrophosphatase (TAP) and polynucleotide kinase (PNK) before ligating 5' adaptors. Adaptor-flanked RNA fragments are enriched through another streptavidin pull-down process before reverse transcription and PCR amplification. Resultant cDNA strands are then sequenced from the 3' end.
Pros:
- Maps RNA pol II pausing sites with base-pair resolution
- Separate run-on reactions limit the addition of nucleotides other than the provided biotin-NTP
- Multiple biotin enrichment steps pre-PCR
- Initiation sites can be mapped using PRO-cap
Cons:
- Unable to detect arrested or backtracked RNA pol II complexes1
- Limited to in-vitro reactions
- PRO-Seq: Kwak H., Fuda N. J., Core L. J. and Lis J. T. (2013) Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950-953
- 1. Weber C. M., Ramachandran S. and Henikoff S. (2014) Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell 53: 819-830
- Wang I. X., Core L. J., Kwak H., Brady L., Bruzel A., et al. (2014) RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep 6: 906-915
- Samarakkody A., Abbas A., Scheidegger A., Warns J., Nnoli O., et al. (2015) RNA polymerase II pausing can be retained or acquired during activation of genes involved in the epithelial to mesenchymal transition. Nucleic Acids Res 43: 3938-3949
- Danko C. G., Hyland S. L., Core L. J., Martins A. L., Waters C. T., et al. (2015) Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods 12: 433-438
- Core L. J., Martins A. L., Danko C. G., Waters C. T., Siepel A., et al. (2014) Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 46: 1311-1320
- Pagano J. M., Kwak H., Waters C. T., Sprouse R. O., White B. S., et al. (2014) Defining NELF-E RNA binding in HIV-1 and promoter-proximal pause regions. PLoS Genet 10: e1004090