MeDIP-Seq/DIP-Seq/hMeDIP-Seq
MeDIP-Seq/DIP-Seq/hMeDIP-Seq
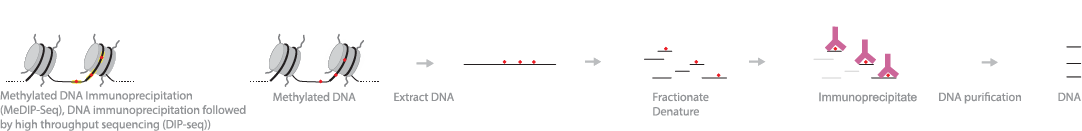
Methylated DNA immunoprecipitation sequencing (MeDIP-Seq), DNA immunoprecipitation sequencing (DIP-Seq) or Hydroxymethylated DNA immunoprecipitation sequencing (hMeDIP-Seq), is commonly used to study 5mC or 5hmC modification. Specific antibodies can be used to study cytosine modifications. If using 5mC-specific antibodies, methylated DNA is isolated from genomic DNA via immunoprecipitation. Anti-5mC antibodies are incubated with fragmented genomic DNA and precipitated, followed by DNA purification and sequencing. Deep sequencing provides greater genome coverage, representing the majority of immunoprecipitated methylated DNA.
- MeDIP-Seq: Weber M., Davies J. J., Wittig D., Oakeley E. J., Haase M., et al. (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853-862
- DIP-Seq: Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A. C., et al. (2013) Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153: 692-706
- hMeDIP-Seq: Jin C., Lu Y., Jelinek J., Liang S., Estecio M. R., et al. (2014) TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res 42: 6956-6971
- Lee H. J., Lowdon R. F., Maricque B., Zhang B., Stevens M., et al. (2015) Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat Commun 6: 6315
- Martin T. C., Yet I., Tsai P. C. and Bell J. T. (2015) coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics 16: 131
- Elliott G., Hong C., Xing X., Zhou X., Li D., et al. (2015) Intermediate DNA methylation is a conserved signature of genome regulation. Nat Commun 6: 6363
- Gascard P., Bilenky M., Sigaroudinia M., Zhao J., Li L., et al. (2015) Epigenetic and transcriptional determinants of the human breast. Nat Commun 6: 6351
- Bose R., Spulber S., Kilian P., Heldring N., Lonnerberg P., et al. (2015) Tet3 mediates stable glucocorticoid-induced alterations in DNA methylation and Dnmt3a/Dkk1 expression in neural progenitors. Cell Death Dis 6: e1793