MBDCap-Seq/MethylCap-Seq/MBD-Seq/MBDCap/MiGS
MBDCap-Seq/MethylCap-Seq/MBD-Seq/MBDCap/MiGS
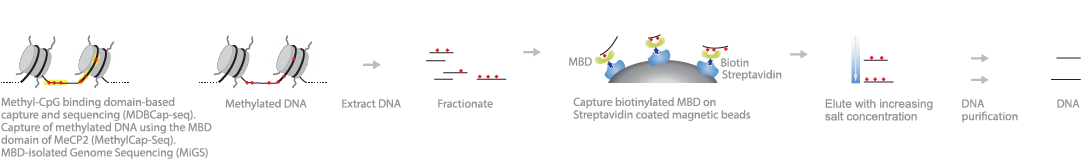
Methyl-CpG binding domain-based capture and sequencing (MethylCap , MBD-Seq , MBDCap-Seq , MBDCap) or MBD-isolated genome sequencing (MiGS ), uses proteins to capture methylated DNA in the genome. Genomic DNA is first sonicated and incubated with tagged MBD proteins that can bind methylated cytosines. The protein-DNA complex is then precipitated with antibody-conjugated beads that are specific to the protein tag. Deep sequencing provides greater genome coverage, representing the majority of MBD-bound methylated DNA.
- Methyl-Cap: Bock C., Tomazou E. M., Brinkman A. B., Muller F., Simmer F., et al. (2010) Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 28: 1106-1114
- Methyl-Cap: Brinkman A. B., Simmer F., Ma K., Kaan A., Zhu J., et al. (2010) Whole-genome DNA methylation profiling using MethylCap-seq. Methods 52: 232-236
- MBD-Seq: Nair S. S., Coolen M. W., Stirzaker C., Song J. Z., Statham A. L., et al. (2011) Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics 6: 34-44
- MBDCap-Seq: de Assis S., Warri A., Cruz M. I., Laja O., Tian Y., et al. (2012) High-fat or ethinyl-oestradiol intake during pregnancy increases mammary cancer risk in several generations of offspring. Nat Commun 3: 1053
- MBDCap: Rauch T. A., Zhong X., Wu X., Wang M., Kernstine K. H., et al. (2008) High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A 105: 252-257
- MBDCap: Rauch T. A. and Pfeifer G. P. (2009) The MIRA method for DNA methylation analysis. Methods Mol Biol 507: 65-75
- MiGS: Serre D., Lee B. H. and Ting A. H. (2010) MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res 38: 391-399
- Bose R., Spulber S., Kilian P., Heldring N., Lonnerberg P., et al. (2015) Tet3 mediates stable glucocorticoid-induced alterations in DNA methylation and Dnmt3a/Dkk1 expression in neural progenitors. Cell Death Dis 6: e1793
- Carrillo J. A., He Y., Luo J., Menendez K. R., Tablante N. L., et al. (2015) Methylome Analysis in Chickens Immunized with Infectious Laryngotracheitis Vaccine. PLoS One 10: e0100476
- Nishikawa K., Iwamoto Y., Kobayashi Y., Katsuoka F., Kawaguchi S., et al. (2015) DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat Med 21: 281-287
- Stirzaker C., Zotenko E., Song J. Z., Qu W., Nair S. S., et al. (2015) Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat Commun 6: 5899
- Yang B. H., Floess S., Hagemann S., Deyneko I. V., Groebe L., et al. (2015) Development of a unique epigenetic signature during in vivo Th17 differentiation. Nucleic Acids Res 43: 1537-1548