药物反应的RNA生物标志物分析
Resources for Adoption of RNA-Seq
RNA sequencing (RNA-Seq) is increasingly being utilized to discover and profile RNA-based drug response biomarkers with the aim of improving the efficiency and success rate of the drug development process. While a number of technologies have been used for this application, the capabilities of RNA-Seq promise to be of particular benefit 1,2,3. Consequently, there is a growing need to make RNA sequencing-based workflow solutions for this application more accessible to a broader range of potential users, including those without prior experience with next-generation sequencing (NGS).
Towards that end, the resources below are designed for users of any level of NGS experience who are considering adopting this application. They contain information that we have found to be particularly helpful across multiple stages of the adoption process, from understanding the steps of an RNA-Seq workflow, to matching configuration options to specific program requirements, to preparing a plan for rapid navigation through the implementation process.

Application
Overview
An introduction to RNA-Seq drug response biomarker discovery and profiling.
Workflow
Introduction
Key considerations, requirements and recommended components for multiple application use-cases.
Best
Practices
"How-to" guidance to facilitate workflow implementation.
Start-up
Advice
Tips from fellow application users and Illumina experts on how to get up and running quickly and smoothly.
Analysis Pipeline
Review
A screenshot-based walk-through from raw data through outputs needed to inform candidate assessment and prioritization.
Section 1: Application Overview
This section provides an overview of RNA-based drug response biomarker discovery and profiling. It includes a review of common methods for this application, including quantitative PCR (qPCR) and gene expression arrays, and the respective strengths and limitations of each. It also reviews the benefits provided by NGS-based workflows and practical considerations about implementation in drug development programs.
Access PDF
Section 2: Workflow Introduction
This section introduces our recommended RNA-Seq workflows for discovering and profiling drug response RNA biomarkers, and outlines the process, from starting total RNA sample through analyzing data.
At each step, the following will be included:
- A high-level description of every step of the process
- Key points to consider when selecting a solution
- Outline of recommended solution(s)

| Step | Library prep | Sequencing | Feature detection | Biomarker candidate ID | Filtering / prioritization |
|---|---|---|---|---|---|
| Requirements |
|
|
|
|
|
| Component |
|
|
|
| Step | Library prep | Sequencing | Biomarker detection |
|---|---|---|---|
| Requirements addressed |
|
|
|
| Component |
|
|
Section 3: Best Practices
This section outlines sequencing-related design parameters that will need to be addressed ahead of planning your study. Included are considerations pertaining to read length, read depth, sequencer output modes, and other variables that should be considered to match the requirements of your program. Also captured are practical considerations related to how transitioning to RNA-Seq from platforms such as quantitative polymerase chain reaction (qPCR) and gene expression (GEX) arrays may affect day-to-day operations, and how you might best prepare.
Access PDF
Section 4: Start-up Advice
Experts across multiple functional areas, as well as users within the pharmaceutical industry currently running this application, offer advice to new users.
Access PDF
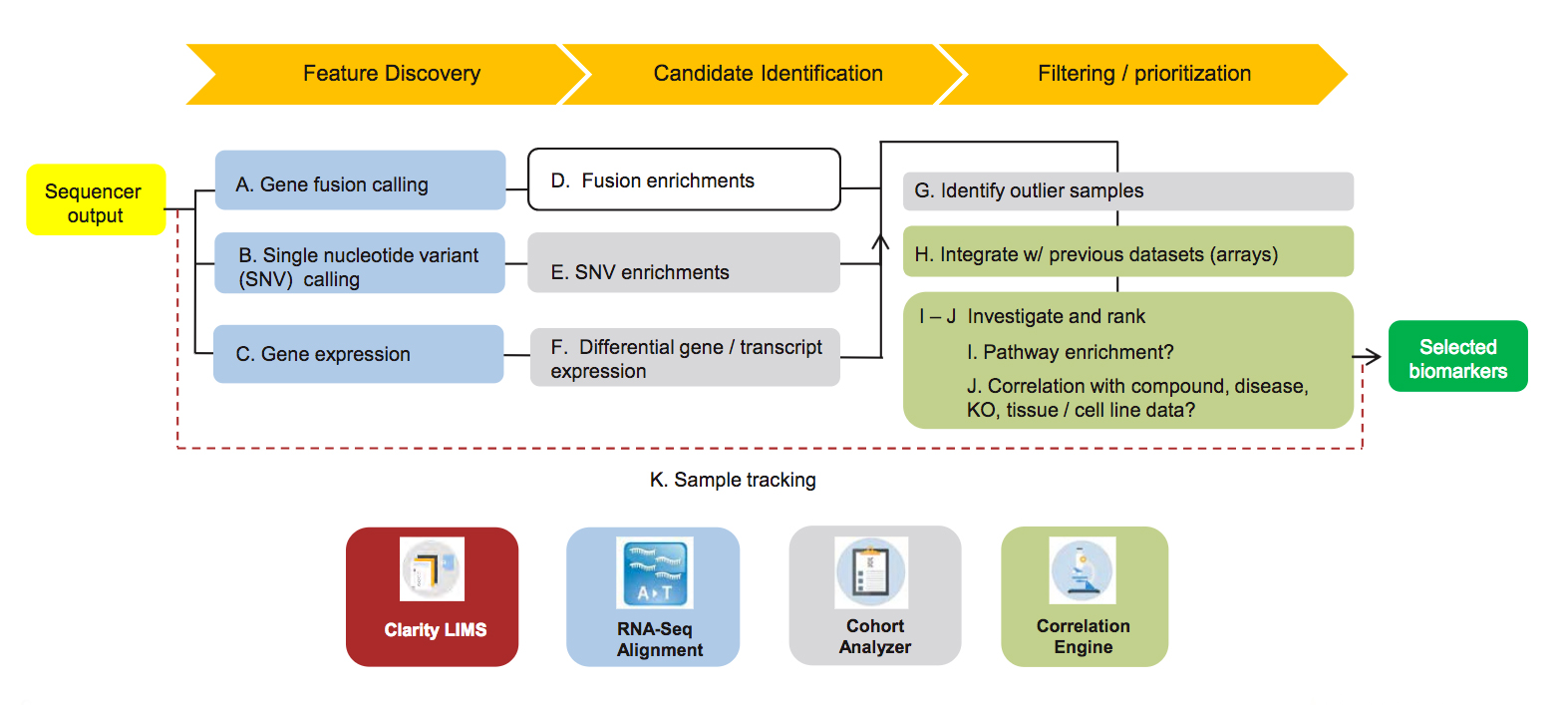
Section 5: Analysis Pipeline Review
Data analysis has historically been one of the most challenging barriers to the adoption of NGS workflows. This has been due, in part, to uncertainty about whether the desired endpoint for a particular application can be reached, what that process entails, and what level of expertise is required. This section provides a holistic view of our recommended analysis pipeline for this application, broken down into feature discovery, identification of RNA biomarker candidates, and biomarker filtering and prioritization. For each work stream within the broader pipeline, a step-by-step, screen shot-based walk-through of the Illumina solution is provided.
View Illumina Pipeline Solution
Access PDF
References
- Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS ONE. 2014;9(1):e78644. doi:10.1371/journal.pone.0078644.
- Atak ZK, Gianfelici V, Hulselmans G, et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013;9(12):e1003997.
- Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129.